Medical imaging is now used extensively for accurate diagnosis, real-time intervention, and precision medicine. Our work has been focused on fast, high-resolution imaging augmented with functional and metabolic mapping. For example, dynamic 4D MRI under free breathing has been developed using PNCRNNs, with the reconstruction time as fast as 50 bin per second, offering substantial improvement of the image quality for dynamic golden-angle radial imaging of the abdomen. Novel imaging techniques have been developed for high-resolution whole-brain neurometabolic mapping, shedding light on in vivo molecular biomarkers for various brain diseases including stroke, brain tumor, and neurodegenerative disorders. A model-based deep learning architecture, referred to as MoDL-QSM, was developed with improved accuracy for quantifying tissue susceptibility for precise electrode implantation for deep-brain stimulation. To meet the demands for automated identification and localization of the vertebrae, pancreas, colon, lung nodules and other organs in low-dose CT, algorithms integrating local image details and global image patterns have been proposed. To extend the clinical use of nonlinear regression in medical imaging and image analysis, a deep negative correlation learning framework has been proposed to yield a deep regression of ensemble where each base model is both accurate and generalisable.
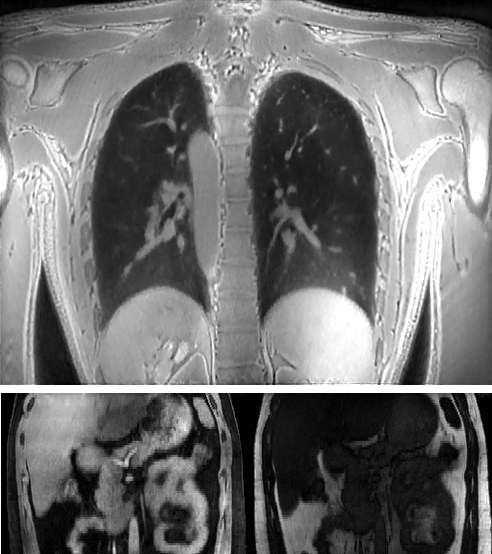
Dynamic 4D MRI of the abdomen
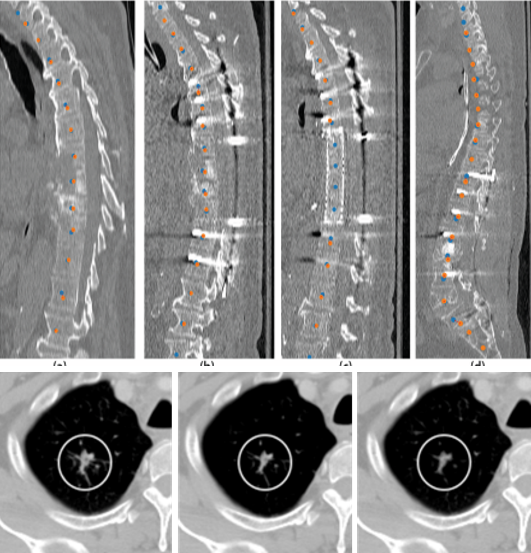
Vertebrae identification and localization
Selected Publications:
1. Zhang Y, She H, and Du YP. Dynamic MRI of the
abdomen using parallel non-cartesian convolutional recurrent neural networks.
Magnetic Resonance in Medicine, 2021, 86:964-973.
2. Li Y, Wang T, Zhang T, et al. Fast high-resolution
metabolic imaging of acute stroke with 3D magnetic resonance spectroscopy.
Brain, 2020, 143(11): 3225-3233.
3. Feng R, Zhao J, Liu C, Zhang Y, and Wei H.
MoDL-QSM: Model-based deep learning for quantitative susceptibility mapping.
Neuroimage, 2021, 204: 118376.
4.
Chen Y, Gao Y, Li K, Zhao L, and Zhao J. Vertebrae identification and
localization utilizing fully convolutional networks and a hidden Markov model.
IEEE Transactions on Medical Imaging, 2020, 39(2): 387-399.
5. Zhang L, Shi Z, Cheng M, et al. Nonlinear
regression via deep negative correlation learning. IEEE Transactions on Pattern
Analysis and Machine Intelligence, 2021, 43(3): 982-998.